I. The Integration of Biology and Language Models
A. Complexity of Biology
Life on Earth originated from chemical reactions 3.5 billion years ago, giving rise to core molecules such as RNA, proteins, and DNA, as well as ribosomes. Proteins, with their myriad of magical functions, are the foundation of life. However, the language of biology is difficult to understand, and current computational tools struggle to delve deeply into research.
B. The Birth of ESM3
ESM3 is a pioneering life sciences language model that takes a step towards a future where biology is programmable, akin to engineering structures, machines, microchips, and writing computer programs.
II. Introduction to the ESM3 Model
A. Training Data and Scale
ESM3 is trained on one of the world's highest-throughput GPU clusters, covering billions of proteins on Earth, including those from extreme environments such as the Amazon rainforest, the deep sea, hydrothermal vents, and microorganisms in the soil. It is a cutting-edge generative biology model with 98B parameters and computational power exceeding 1x10^24 FLOPS.
B. Multimodal Reasoning Capabilities
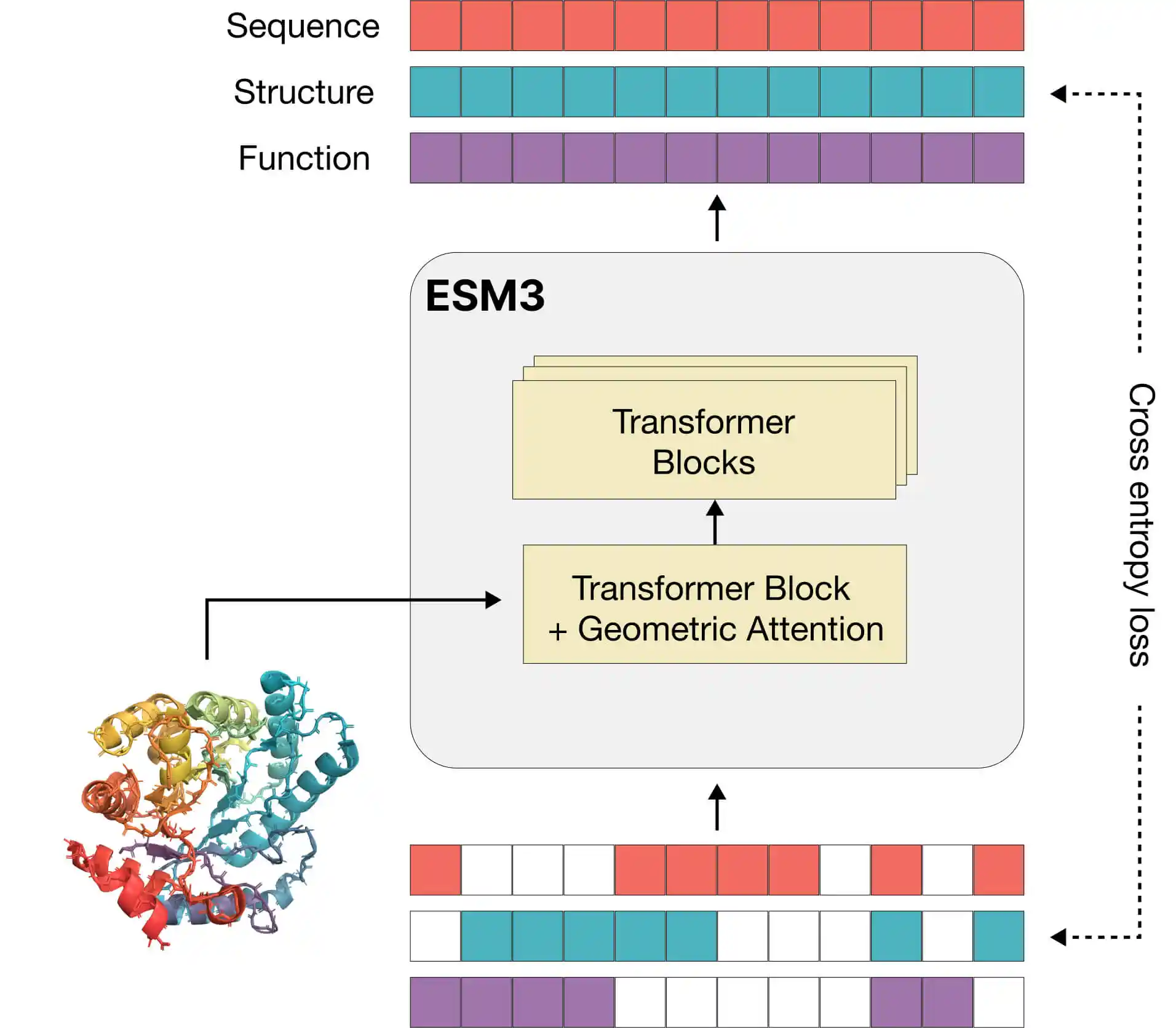
Reasoning about proteins: ESM3 is a multi-track transformer capable of reasoning about protein sequences, structures, and functions simultaneously. It translates three-dimensional structures and functions into a discrete alphabet, allowing the vocabulary to connect sequences, structures, and functions within the same language model.
Training approach: For each protein, its sequence, structure, and function are extracted, tokenized, and partially masked. ESM3's task is to predict the masked positions using masked language modeling objectives, thereby learning the connections between sequences, structures, and functions, simulating evolution.
Generating proteins: It can start from a completely masked token set, iteratively unmask to generate new proteins. Since sequences, structures, and functions are all masked and predicted during training, they can be generated in all three modalities. Scientists can guide the generation process with prompts for various applications such as medical research, biology, and clean energy.
C. Emergence and Enhancement of Capabilities
Emergence of capabilities: As the model size increases, ESM3's ability to solve challenging protein design tasks, such as atomic coordination tasks, gradually emerges, and its ability to solve these tasks improves with scale.
Self-enhancement: ESM3 can self-improve using alignment methods similar to human feedback reinforcement learning (RLHF), providing feedback based on the quality of its own generation. Feedback from laboratory experiments or existing experimental data can also be used to align generated results with successful biological cases.
III. Experiment Simulating 50 Million Years of Evolution
A. Green Fluorescent Protein (GFP)
GFP and its fluorescent protein family are beautiful and important proteins in nature, which can be used to observe proteins inside cells. Its unique fluorescent chromophore mechanism allows it to absorb short-wavelength photons of one color and emit photons of a different color and longer wavelength. Scientists have discovered many GFP variants, and most functional GFP variations come from nature rather than protein engineering.
B. ESM3's Simulation Experiment
Generating new GFP: Based on the structure of some core residues of natural GFP, ESM3 generates new GFP candidates through chain of thought reasoning. In the first experiment, 96 generated proteins were tested, and some proteins with fluorescent functions were found, one of which is very different from any protein in nature. Continuing from the sequence of this protein (B8), another 96 proteins were generated and tested, discovering several proteins with brightness similar to natural GFP, including esmGFP. esmGFP has 96 mutation differences from the closest fluorescent protein in nature (58% sequence similarity in 229 amino acids).
Evolution simulation: Although protein language models are not subject to evolutionary constraints like nature, to complete the training task of predicting the next masked token, ESM3 must learn the way evolution occurs in protein space, thus it can be considered an evolution simulator. Analysis of esmGFP estimates that it is equivalent to more than 50 million years of natural evolution in the evolution simulator.
IV. Responsible Development
A. Development Framework
EvolutionaryScale is a public benefit corporation that has established a responsible development framework, including core principles such as communicating the benefits and risks of research, strictly assessing model risks before public deployment, adopting risk mitigation strategies and preventive measures, and collaborating with stakeholders.
B. Open Model
The ESM project has been committed to open science, releasing code and models. This time, the weights and code of the ESM3 1.4B open model will be released to accelerate research and empower the scientific community.
V. Future Development Directions
A. Assisting Scientists in Exploration
ESM3 is a tool for scientists, and its API and open models allow scientists to explore the frontiers of protein design and synthetic biology, inventing new solutions for some of the world's most important problems. Priority will be given to beta access to the API based on the potential to expand the scientific knowledge frontier and create new tools that benefit the world.
B. Application in Drug Design
A specialized version of ESM3 is being developed to unlock applications at the forefront of drug design, helping scientists create new drugs. ESM3 is just the first step on the roadmap of programmable biology, with the future being more modal models that learn biological data from molecules to cells, contributing to human understanding and programming of biology, and building a better world.

